Gene expression and development
The embryonic genome of many vertebrate and invertebrate species is transcriptionally quiescent during the earliest stages of development. In our model system for vertebrate embryonic development, Xenopus laevis, the embryonic genome is activated after 12 cell divisions (4000 cells), a process referred to as the mid-blastula transition (MBT). At the MBT many genes are transiently derepressed, regardless of the normal stage or cell type of expression. At the onset of gastrulation - shortly thereafter - spatially restricted expression patterns are established by targeted transcription activation and repression events.
|
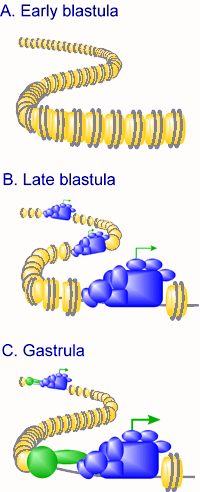
Transcriptional regulation during early embryogenesis in Xenopus. The embryonic genome is transcriptionally quiescent before the mid-blastula transition (MBT), due to a repressive chromatin structure (symbolized in panel A in the figure to the right by a high density of nucleosomes), and constraints on the transcription machinery such as low rate-limiting levels of TATA binding protein (TBP), cytoplasmic retention of transcriptional regulators, and constraints on transcriptional activation. In late blastula embryos, chromatin is less repressive toward transcription (symbolized in panel B by lower density of nucleosomes) and transcription of a subset of genes is facilitated by the regulated translation of TBP mRNA. Generally transcription levels are low, and are mediated by the basal transcription machinery, with constraints on activator function still in place. The general transcription machinery is depicted at "open" spots of the chromatin, with the transcription start site depicted with an arrow. During gastrulation chromatin becomes more repressive towards transcription due to incorporation of linker histone H1 into chromatin and a more prominent role of histone deacetylases. A more prominent role for targeted, gene-selective activation and repression events is observed, symbolized by the presence of additional proteins (circle and oval) in the vicinity of the general transcription machinery. As a consequence, some genes are induced to high levels whereas some of the genes that were transcribed initially are repressed by the time of gastrulation (Veenstra, 2002). |
|
The molecular mechanisms underlying this regulation are largely unknown, but are known to involve the counteracting effects of chromatin-mediated repression of basal transcription, and global as well as gene-specific activation overruling transcriptional repression. Current data suggest that chromatin-mediated repression of basal transcription plays a role in the transcriptional quiescence before the MBT, whereas developmental regulation of general transcription factors such as TATA-binding protein (TBP) and TBP-like factor (TLF) have been implicated in activation of the embryonic genome at the MBT.
There are several important reasons to study early embryonic gene regulation. Firstly, studying the global regulation of the embryonic genome provides an opportunity to gain insight into the molecular mechanisms that govern developmental control of gene expression. Secondly, transcriptional activation of the embryonic genome is pivotal to the regulatory hierarchy of genes during the earliest stages of embryogenesis. Genes in this hierarchy include genes essential for mesoderm and neural induction, and patterning. Xenopus is an ideal model system to study these processes, which is essential to further our knowledge of both normal development and disease such as congenital malformation or abnormal growth and differentiation.
